Literature Review Sample Work
Advancements in MALDI-TOF MS for Quick Identification of Microbial Pathogens: A Comprehensive Synopsis
Info: 2935 words Sample Literature Review
Published: 27th OCTOBER 2023
Tagged: Biology & Lifescience
Abstract
Every year, plant diseases caused by plant pathogens significantly impair food yield, resulting in large economic losses throughout the world. Accurate detection and Identification of plant pathogens is critical for plant pathogen diagnostics and, therefore, plant disease management. Diagnostics and disease management methods necessitate the simultaneous Problem Identification and quantification of a diverse variety of pathogenic and nonpathogenic microorganisms. Over the last decade, the fast development of matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) methods for microbe characterization has resulted in much-enhanced detection and Identification of microorganisms. MALDI-TOF MS is used in the biological sciences to evaluate particular peptides or proteins immediately desorbed from intact bacteria, fungal spores, nematodes, and other microorganisms. Taxonomic Identification of microorganisms by MALDI-TOF MS is based on the capacity to record biomarker ions in a broad m/z range that are unique to and typical of particular bacteria. Recent advancements in mass spectrometry have sparked increased interest in the study of increasingly complex microbial communities. Such research is in its early stages, but it has the potential to provide light not just on interactions between microbes and their host plants but also on interactions between diverse microbial species living in conjunction with plants. The mass spectrometry community has recently attempted to make data from large-scale mass spectrometry studies publicly available in the form of a consolidated repository. With such a resource, MALDITOF MS might be used as a universal approach for detecting plant pathogens and non-pathogens. The effects of experimental synopsis settings are well established; repeatable spectra may be acquired using computerized database searches, and genus, species, or strain can quickly identify microorganisms.
MALDI-TOF MS as an alternative to molecular approaches for the quick Identification of plant microbes
Pathogenic bacteria, fungi, nematodes, and viruses constitute a persistent hazard to plant propagation material production. Reliable and appropriate technologies for screening numerous pathogens are critical for implementing successful disease control programs. Thus, early detection and Identification of plant-associated microbes is an essential component of successful disease control and is especially crucial in the context of foreign plant material importation. Rapid Identification of plant-associated bacteria allows for the application of suitable management measures prior to disease development or spread. Because most plant-based foods have a short shelf life, it is critical that any potentially contaminated portion be identified as soon as feasible and as accurately as possible to minimize delays and costly losses. Traditionally, the most common methods for identifying plant pathogens depended on which are complex and needed a high level of taxonomic expertise, professional training, and experience [1]. The overall identification time, including traditional culture time (morphology of cell colonies and slide culture), is about 2-14 days, depending on the varying growth rates of various species; this frequently delays illness management. However, in recent years, several techniques for plant pathogen diagnosis have been introduced, including the use of monoclonal antibodies and enzyme-linked immunosorbent assay (ELISA), which have significantly increased the speed of in vivo detection of pathogenic antigens and DNA-based techniques, such as polymerase chain reaction (PCR), which allow regions of the pathogen's genome to be amplified several millionfold.
PCR, which was introduced in the mid-1980s, is currently utilized to study plant-microbe interactions [3]. Its application in horticulture and agriculture is quickly growing, and the availability of such diagnostic assays benefits a variety of fields. With many nations' borders opening and growing free-trade agreements, fast testing for probable contamination with quarantine organisms is critical. Furthermore, the need for quick and economical pathogenic detection tests is growing in order to enable prompt application of control measures. However, certain requirements must be followed before novel detection methods may be used in academic and scientific settings. These rules can be separated into two categories: technical and economic requirements. While technical requirements are necessary for the creation of any effective diagnostic procedure, economic factors are also significant.
This review paper highlights the challenges and accomplishments in the development and deployment of specialized MALDI-TOF MS in plant pathology. Figure 1 displays the MALDI-TOF MS methodologies used to characterize plant-associated bacteria. Fungi, bacteria, nematodes, viruses, and virus-like organisms are the four basic types of plant-associated microorganisms. The Identification of plant-associated microorganisms (pathogens/non-pathogens) in the first three groups using MALDI-TOF MS will be explored in this study.
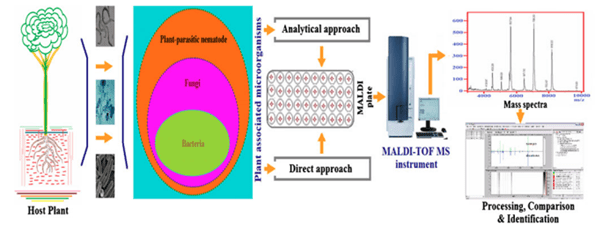
Fig. 1 Pictorial illustration of strategies for rapid characterization of plant-associated microorganisms by MALDI-TOF MS and computational data processing (Sechul Chun et al. 2022)
MALDI-TOF mass spectrometry and plant microorganisms
- Bacterial Identification using mass spectrometry
- characterization;
- classification; and
- nomenclature.
- absorption at the laser wavelength;
- ability to dissolve or co-crystallize with the sample;
- low volatility;
- ability to suppress analyte decomposition and
- ability to promote ionization of the analytes.
- Larone DH (1995) Washington DC: ASM Press, 190–192
- Babalola OO, Kirby BM, Le Roes-Hill M, Cook AE, Cary SC, Burton SG, Cowan DA (2009) Environ Microbiol 11:566–576
- Babalola OO (2003) Afr J Biotechnol 2:710–713
- Lopez MM, Bertolini E, Marco-Noales E, Llop P, Cambra M (2006) Norfolk, UK: Horizon Bioscience, 1–46
- Pan YL, Chow NH, Chang TC, Chang HC (2011) Diagn Microbiol Infect Dis 70:344–354
- Hillenkamp F, Karas M (2000) Int J Mass Spectrom 200:71–77
- Anhalt JP, Fenselau C (1975) Anal Chem 47:219–225
- Dworzanski JP, Snyder AP (2005) Expert Rev Proteomics 2:863– 878
- Nugen SR, Baeumner AJ (2008) Anal Bioanal Chem 391:451–454
- Naja G, Bouvrette P, Hrapovich S, Liu Y, Luong JHT (2007) J Raman Spectrosc 38:1383–1389
- Yang H, Qu LW, Lin Y, Jiang XP, Sun YP (2007) J Biomed Nanotechnol 3:131–138
- Zhang HQ, Zhao Q, Li XF, Le XC (2007) Analyst 132:724–737 Tan KC, Ipcho SV, Trengove RD, Oliver RP, Solomon PS (2009) Mol Plant Pathol 10:703–715
- De Wit PJGM, Buurlage MB, Hammond KE (1986) Physiol Plant Pathol 29:159–172
- Ellis JG, Rafiqi M, Gan P, Chakrabarti A, Dodds PN (2009) Curr Opin Plant Biol 12:399–405
- Rep M, van der Does HC, Meijer M, van Wijk R, Houterman PM, Dekker HL, de Koster CG, Cornelissen BJ (2004) Mol Microbiol 53:1373–1383
- Chou PH, Chen SH, Liao HK, Lin PC, Her GR, Lai ACY, Chen JH, Lin CC, Chen YJ (2005) Anal Chem 77:5990–5997
- Teng CH, Ho KC, Lin YS, Chen YC (2004) Anal Chem 76:4337– 4342
- Chen WJ, Tsai PJ, Chen YC (2008) Anal Chem 80:9612–9621
- Armstrong DW, Zhang LK, He L, Gross ML (2001) Anal Chem 73:3679–3686
- Crank JA, Armstrong DW (2009) J Am Soc Mass Spectrom 20:1790–1800
It has long been understood that studying plant diseases would be impossible without a comprehensive definition of the microorganisms. In addition to dangerous bacteria, roots contain a variety of helpful bacterial species known as endophytic bacteria. As a result, tremendous effort has been expended since its start to improve current techniques for microbial characterization continuously. MALDI-TOF MS was developed two decades ago to detect biomarkers in target bacterial samples. MALDI-TOF MS has been successfully applied to a wide range of microbial applications, including bacterial RNA and DNA analysis, detection of recombinant proteins, characterization of unknown proteins, bacterial proteomics, detection of virulence markers, and very fast bacterial characterization at the genus, species, and strain levels.
Bacteria have a diverse set of proteins that allow them to proliferate in the plant and effectively infect it. Bacterial taxonomy may be divided into three parts:
The usual strategy for microbiological Identification has been phenotypic characterization. However, during the last 50 years, the emphasis on bacterial characterization has changed from phenotyping to genotyping.
MALDI-TOF MS is increasingly being used in microbiology taxonomic difficulties, such as detecting cryptic species in large batches of related isolates [17]. However, it is currently not possible to determine conclusive similarity values for mass spectral signals acquired from non-specific isolates, i.e., a species-separating threshold. This is not totally unexpected, given that the same principles for designating microbial species do not apply to all taxa. Furthermore, past biases have characterized live microorganisms using criteria that are not always congruent with contemporary ideas on how bacterial species should be described.
Extensive research has also been conducted to identify plant-beneficial bacteria and their application in agriculture to lessen the impact of pathogens that cause a range of plant illnesses. Plant growth-promoting rhizobacteria (PGPR) are a diverse collection of bacteria found in the plant rhizosphere that contribute to enhanced yields of crops, vegetables, and other economically important plants. PGPR stimulates beneficial activity that is responsible for the manufacture of phytohormones and volatile organic compounds that are accessible as plant nutrients, and there have been multiple findings that imply a reduction of phytopathogenic soil bacteria, fungi, viruses, and nematodes [20].
For applications that do not need quick or real-time analysis, the use of chromatographic techniques prior to mass analysis has also been considered. On-line and off-line chromatographic methods have been integrated with MS. Typical of this work is that published by Fox et al., who used gas chromatography-MS to separate two closely related Bacillus spp., Bacillus anthracis and B. cereus, which are difficult to distinguish phenotypically or genotypically [21]. Cain et al. used chromatography (off-line) and MALDI-TOF MS to differentiate bacteria based on protein fractions extracted from damaged cells [22]. This research was significant for using proteins as biomarkers rather than smaller chemicals.
MALDI mass spectrometry-based bacterial analysis has matured in recent years, and numerous excellent papers that clarify the many methodologies involved in bacterial characterization have been authored using reported data [26]. These biorecognition technologies can identify infections quickly and accurately. Within minutes, nanoparticles are coupled with biomolecules such as antibodies, antibiotics, adhesion molecules, and complementary DNA sequences for pathogen identification. Device miniaturization has resulted in scientific laboratories on tiny chips that can detect diseases using portable, hand-held biosensors [27]. The nanoparticle's versatile chemistry, unusual optical characteristics, and strong ferromagnetic responses [28, 29] have led to its employment as an efficient tool in several biodetector systems.
Fungus detection using mass spectrometry
Until the early 1990s, most biological research was focused on in vitro examinations of specific components, such as genes and proteins. In the early and mid-1990s, this method switched to in vivo and molecular large-scale research, beginning with structural genomics and transcriptomics studies, then moving on to proteomics and, more recently, metabolomics. All of these represent the methodological foundations of current systems biology [31]. Because no one technique can explain the complexity of living organisms, each approach that contributes must be validated and recognized as part of an interdisciplinary, integrative study at several levels, ranging from the gene to the phenotype via proteins and metabolites. Proteomics science is constantly being renewed, and in the last ten years, there has been a massive increase in the number of new strategies and procedures, as well as techniques, with continuous improvements made at all stages of the procedure, beginning in the laboratory (tissue and cell fractionation, protein extraction, depletion, purification, separation, mass spectrometric analysis) and ending at the computer (algorithms for protein recognition and bioinformatics tools for data analysis).
Wheat leaf rust, caused by the fungus Puccinia triticina, has also been studied using proteomic analysis [37]. Rust infections reduce cereal crop yields by a large amount each year [38]. The proteomes of both the host and the pathogen were examined during illness progression to gain a better understanding of the problem at the molecular level. Using 2DE (with isoelectric focusing, pH 4-8) and mass spectrometric analysis, a susceptible line of wheat infected with a virulent race of leaf rust was compared to mock-inoculated wheat [37]. Schmidt and Kallow used MALDI-TOF MS to identify the closely related indoor wood-decay fungus Serpula lacrymans, S. himantioides, Coniophora puteana, C. marmorata, Antrodia vaillantii, and A. sinuosa [39].
Identification of plant-parasitic nematodes using mass spectrometry
Plant parasitic nematodes, which are also hidden enemies of plants, frequently cause symptoms that are readily misunderstood. These microscopic creatures are cropping system pests rather than single-crop pests, especially in tropical locations where cropping intensity is high, and circumstances support the development of a large nematode population. Parasitic nematodes can significantly impair crop development and output in a variety of commercially important crops [42]. Endoparasitic root nematodes may cause massive crop output losses and have thus been widely investigated. Most economically important plants are vulnerable to attack by one or more plant-parasitic nematode species, and their interactions with the plant host can range from transitory grazing by root hair feeders to the highly complex host-pathogen interactions of endoparasites and gall-inducing nematodes.
Morphologically, nematodes are quite similar. Caenorhabditis elegans and Caenorhabditis briggsae are two free-living worms whose Identification would be difficult for most skilled nematode taxonomists. Despite their physical similarities, a recent analysis of 338 orthologous genes (homologous genes generated from the same gene in the last common ancestor) revealed that the two species diverged 80-110 million years ago [49]. This breaking happened 5-45 million years before the mouse and human lineages separated [50]. While most of us are comfortable making the man-mouse distinction, this example sends a strong warning to those who must identify nematode species. Clearly, cryptic species must exist in the phylum Nematoda, and molecular methods may be the only way to identify them. Protein, lipid, carbohydrate, and DNA analysis, as well as molecular techniques such as protein electrophoresis, restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), dot blot assays, and variation in internally transcribed spacer (ITS) sequences detected by polymerase chain reaction, have all been used to identify nematodes [51, 52].
Mass spectrometric imaging
Mass spectrometry advancements have resulted in new exploratory investigations of molecular interactions in intact tissue, similar to studies undertaken decades ago using intact tissue for metabolic research, but now with exact molecular specificity. MALDI mass spectrometric imaging (MALDI-TOF MSI), in particular, allows the researcher to examine the spatial distribution of sugars, metabolites, and lipids in plant tissues. The capacity to monitor many substances within a cell or tissue at the same time is very appealing. MALDI-TOF MSI has the benefit of not requiring the creation of an antibody linked with a fluorescent or transmission electron microscopy probe, as well as the fact that MALDI-MS equipment is widely used. The MALDI-TOF MSI technique has been utilized effectively in plant biology.
Chemical approach before MALDI-TOF MS
Analytical approach
Several analytical procedures have been employed to identify bacteria in a variety of biological materials [16, 67]. TEM, MALDI-TOF MS, combined nanoparticles/luminescence-based assays [68], and magnetic bead-based immunological assays [69] are among these approaches. Regardless of the detection approach, the nanoparticle must successfully and specifically target a specific bacterium (or, more generally, a biomolecule). This is only possible if its surface is adequately functionalized with substrates capable of selectively and firmly engaging with surface groups on the biomolecule of interest. Antibody-modified nanoparticles have been effectively employed in microbial cell and biomolecule labelling investigations in combination with MALDI-TOF MS [16].
MALDI matrices approach
The mechanism of ion generation in the process of desorption/ionization under the action of laser pulses has been systematically summarized [11]. Thus, the success of MALDI measurement is partly empirical and remains highly dependent on the type of matrix and appropriate target sample preparation prior to MALDI-TOF MS analysis. In MALDI-MS analysis, matrix selection is critical. The following are the characteristics of an ideal matrix:
A matrix can be either solid or liquid. In MALDI analysis, the most widely employed solid matrices are derivatives of weak organic acids, such as -cyano-4-hydroxycinnamic acid (CHCA), sinapinic acid (SA), and 2,5-dihydroxybenzoic acid (2,5-DHB). The use of matrix additives is a straightforward technique to improve the quality of MALDI spectra for certain data analytics, either by suppressing sodium adducts or boosting analytical sensitivity and resolution. Armstrong et al. [75] were the first to employ ionic liquids (ILs) as matrices successfully. Because of their unique features (low volatility and excellent solvents for a wide range of compounds), ILs are well suited for use as MALDI matrices. A second generation of IL matrix for MALDI-TOF MS has recently been created [76].
Determining remarks and future perspectives
Diagnostic laboratories and inspection departments are increasingly looking for new instrumental techniques that allow for reliable Identification, sensitive detection, and precise quantification of possible plant pathogenic and nonpathogenic organisms. The findings discussed in this review show how MALDI-TOF mass spectrometry has been utilized to improve the Identification of plant-associated bacteria. MALDI-TOF MS-based proteomics will play an important role in the Identification, characterization, and improved understanding of the biology of plant-associated microbes such as bacteria, fungi, and nematodes. This low-cost protein mass pattern detection approach is simple to use as a diagnostic tool for detecting microorganisms at the genus, species, and, in some circumstances, subspecies levels.
Conclusion
A topic of great interest that impacts humanity and has major ramifications for agricultural plants, resulting in plant illnesses, has been evaluated using MALDI-TOF MS. The results and advancements made using MALDI-TOF MS for detecting bacterial, fungal, and viral pathogens have been described. The reasons for the poor interest in MALDI technology deployment for precise, quick detection of plant diseases have been examined.
Check out our blog to learn more about Maximizing the Impact of Your PhD Manuscript: Challenges and Publishing Opportunities.
References
Related Services
Our academic writing and marking services can help you!
Study Resources
Free resources to assist you with your university studies!
